
CMU Receives $7.5M in Federal BRAIN Initiative Funding
Grants support creation of new technologies for understanding the brain
Researchers from Carnegie Mellon University's departments of Biological Sciences and Chemistry, Molecular Biosensor and Imaging Center (MBIC) and Pittsburgh Supercomputing Center (PSC) have received close to $7.5 million in new funding from the National Institutes of Health through the federal BRAIN Initiative to support innovative research and develop tools that will rapidly advance brain research.
"Carnegie Mellon's combined expertise in biology, psychology, computer science and engineering has positioned us to be at the forefront of creating new tools and technologies for neuroscience," said Alison Barth, professor of biological sciences, interim director of CMU's BrainHub neuroscience initiative and a member of the Center for the Neural Basis of Cognition. "The federal BRAIN Initiative's support is invaluable to increasing our understanding of how the brain communicates."
High Throughput Approaches for Cell-Specific Synapse Characterization
Marcel Bruchez, director of MBIC and professor of biological sciences and chemistry, and Barth were awarded more than $2 million to develop new tools and methods for high-throughput fluorescence synapse quantitation. They will work with the University of Pittsburgh's Simon Watkins, director of the Center for Biologic Imaging (CBI).
The human brain consists of billions of neurons, each of which passes information through thousands of synapses. Identifying how and where synapses develop under normal conditions and viewing how they change is vital to our understanding of learning, development and disease.
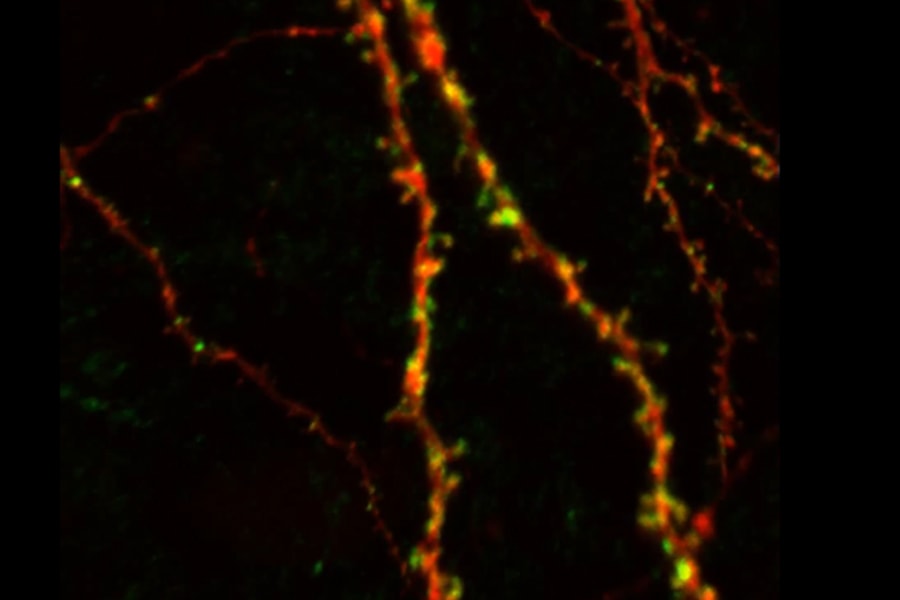
An image shows the ostsynaptic labeling of dendritic spines using fluorogen-activating proteins in a pyramidal neuron in the neocortex.
Fluorescence imaging has made it possible to identify specific cells among the sea of neurons in the brain and monitor how they are connected with one another. Under the new grant, the researchers will optimize reagents that will allow them to detect presynaptic and postsynaptic inputs and quantify synapse distribution between specific cell types in brain tissue. They will also develop software that will automate the analysis of the data, which was previously done by humans. This will allow researchers to count millions of neurons and billions of synapses, whereas previous studies were only able to look at small numbers of cells, or subcellular regions.
Confocal Fluorescence Microscopy Data Repository
Alexander J. Ropelewski, director of the biomedical application group at the PSC, Bruchez and Watkins received more than $5 million under the BRAIN Initiative Cell Census Network to build a confocal fluorescence microscopy data repository that will provide researchers with easy, searchable access to petabytes of neuroscience data.
New imaging tools and technologies, like large-volume confocal fluorescence microscopy, have greatly accelerated neuroscience research in the past five years, allowing researchers to image the whole brain at such a high level of resolution that they can zoom in to the level of a single neuron or synapse. These images, however, contain such a large amount of data — that only a small part of one brain's worth of data can be accessed at a time using a standard desktop computer. Additionally, images are often collected in different ways — at different resolutions, using different methodologies and different orientations. Comparing and combining data from multiple whole brains and datasets requires the power of supercomputing.
The Pittsburgh-based team will bring together MBIC and CBI's expertise in cell imaging and microscopy and pair it with the PSC's long history of experience in biomedical supercomputing to create a system called the Brain Imaging Archive. Researchers will be able to submit their whole brain images, along with metadata about the images, to the archive. There the data will be indexed into a searchable system that can be accessed using the internet. Researchers can search the system to find existing data that will help them narrow down their research targets, making research much more efficient.
Machine Learning Approaches for Electrophysiological Cell Classification
Barth was awarded more than $300,000 to develop a method to identify neural cell types from extracellularly recorded spike trains, a "Rosetta Stone" for decoding brain activity.
In a 500x500 micron column of brain tissue, there can be more than 60 different types of neuronal cells, each with its own properties. Currently researchers gather information from neurons by inserting an electrode into the brain and recording when neurons spike in response to a stimulus. While this reveals valuable information, researchers don't know which cell type they are recording.
Barth believes that researchers could determine cell type based on patterns found in a neuron's spike train. She will partner with researchers in Carnegie Mellon's Machine Learning Department and Language Technology Institute to develop classifiers for neuron types and use machine learning to identify cell type based on the pattern of spikes collected from a neuron.
Previously, Carnegie Mellon researchers received an additional $2.8 million from the BRAIN Initiative. Engineering faculty members Steven Chase and Byron Yu received over $850,000 from the National Science Foundation (NSF) to establish how variability in movement is encoded in the brain and how this variability contributes to learning and performance. Yu also received close to $1 million from the NSF to understand how the sensory environment and state of mind combine to affect perception and interpretation of the world around us. Max G'Sell, assistant professor of statistics received close to $1 million to study brain circuits in a real-world setting.
